Grant: 12-009R
Project Title: Genetic assessment of the distribution of juvenile green turtles from the Florida rookery on southeastern United States foraging grounds
Project Manager: Campbell Nairn
Organization: University of Georgia Research Foundation - Natural Resources (Research and Educational Institute)
Grant Amount: $37,087.00
Completion Date: 2013-06-21
Summary: The presence of three common 480-bp mitochondrial control region haplotypes among Greater Caribbean rookeries limits fine scale resolution of mixed stock analyses. We will confirm that Florida-excluded CM-A1 turtles could be of Mexican origin through mitogenomic sequencing of Rancho Nuevo nesting female CM-A1 samples archived at SWFSC, which will expand baseline rookery data. We will perform a many-to-many mixed stock analysis of approximately 1700 juvenile green turtle foraging aggregation samples based on expanded control region sequences and a subset of mitogenomic haplotypes. The mixed stock analysis will focus on juvenile foraging aggregations that have not been previously characterized or have been characterized based solely on the short haplotypes: Florida Bay, the Mosquito Lagoon/Banana River, and St. Joseph Bay in Florida as well as Texas, North Carolina, and Nicaragua. The Florida samples were all collected during hypothermic stunning events in the winters of 2000-2001 and 2010, which provided a snapshot of turtles spanning the different size classes. These snapshots in time and space will permit testing for temporal variation across the decade time span at St. Joseph Bay and in Texas. The sampling will also provide an opportunity to test whether turtles of different size classes using the same foraging macrohabitat represent cohesive cohorts of similar origins or represent ontogenetic clusters of turtles transitioning through foraging habitats based on their sizes.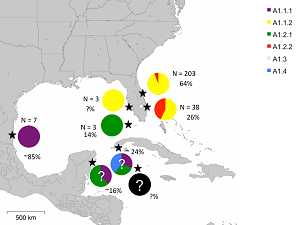 Results: This project confirmed that the Rancho Nuevo nesting females carry haplotype CM-A1.1.1. Haplotype frequencies were significantly different for all foraging aggregations analyzed except the northern Indian River Lagoon complex and North Carolina. The mitogenomic CMA1
marker permitted resolution of the distribution of Florida turtles on Gulf of Mexico
foraging aggregations for the first time. Despite the high frequency of CM-A1 in the
Texas foraging aggregation, only five individuals out of 221 were assigned to the Florida
rookery. Florida contributions to the St. Joseph Bay foraging aggregation as inferred from
CM-A1 mitogenomic haplotype distributions were slightly higher (22 of 174), but still
suggest primary contributions from the Mexican management units. Florida contributions
to the foraging aggregations on the east coast of the United States were the highest
confirmed in the study (~26% and ~37% in the Indian River Lagoon and North Carolina
aggregations, respectively). Even in these cases, contributions from combined Mexicanand Costa Rican rookeries accounted for most of the turtles. These results suggest that
other major foraging habitat for green turtles originating from the Florida rookeries
remain to be identified and characterized via mitogenomic sequencing. Please see the
attached Appendix for detailed descriptions of lab methodology, data analysis methods,
and results.
Results: This project confirmed that the Rancho Nuevo nesting females carry haplotype CM-A1.1.1. Haplotype frequencies were significantly different for all foraging aggregations analyzed except the northern Indian River Lagoon complex and North Carolina. The mitogenomic CMA1
marker permitted resolution of the distribution of Florida turtles on Gulf of Mexico
foraging aggregations for the first time. Despite the high frequency of CM-A1 in the
Texas foraging aggregation, only five individuals out of 221 were assigned to the Florida
rookery. Florida contributions to the St. Joseph Bay foraging aggregation as inferred from
CM-A1 mitogenomic haplotype distributions were slightly higher (22 of 174), but still
suggest primary contributions from the Mexican management units. Florida contributions
to the foraging aggregations on the east coast of the United States were the highest
confirmed in the study (~26% and ~37% in the Indian River Lagoon and North Carolina
aggregations, respectively). Even in these cases, contributions from combined Mexicanand Costa Rican rookeries accounted for most of the turtles. These results suggest that
other major foraging habitat for green turtles originating from the Florida rookeries
remain to be identified and characterized via mitogenomic sequencing. Please see the
attached Appendix for detailed descriptions of lab methodology, data analysis methods,
and results.
Figure at right: Distribution of CM-A1 subhaplotypes across rookeries. Frequencies of CM-A1 relative to other control region haplotypes are indicated by percentages. Mitogenomic
haplotype frequencies are speculative for Quintana Roo, Mexico (based on Abreu-
Grobois et al. 2008) and Guanahacabibes Peninsula, Cuba (based on Azanza-Ricardo
2009) and completely unknown for the Cayman rookery. Test samples from the
southwestern Florida rookery confirm that the CM-A1.1 turtles are of the same lineage as
those nesting on the east coast of Florida rather than the lineage nesting in the western
Gulf of Mexico, but additional sequencing is required to determine the relationship of this
rookery to those on the Atlantic coast.