Grant: 09-012R
Project Title: Genetic Population Structure of Loggerheads Nesting in Atlantic Coastal Florida: How Many Subpopulations and Where are the Boundaries?
Project Manager: Campbell Nairn
Organization: University of Georgia Research Foundation - Natural Resources (Research and Educational Institute)
Grant Amount: $17,600.00
Completion Date: 2010-06-17
Summary: The research project addresses outstanding questions regarding genetic population structure among loggerhead subpopulations nesting on peninsular Florida beaches. Population structure analyses conducted with samples collected in 2006 and 2007 suggest that current management units do not fully reflect genetic and demographic partitioning among Florida loggerhead nesting beaches. Genetic analyses conducted to date have been hampered by small sample sizes and/or insufficient geographic coverage to make robust inferences about the integrity of inferred management units or the precise boundaries between them. Collaborators have dramatically increased sampling effort during the 2008 nesting season to address: 1) whether the current recovery unit designations should be refined, 2) how haplotype frequencies transition from beach to beach, 2) haplotype frequency changes across and within seasons in transitional areas, 3) connectivity of northern subpopulation nesting beaches through dispersal of female nesting effort.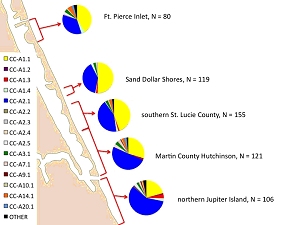 Results: Genetic studies have provided invaluable information regarding population subdivision, particularly cryptic structure present in highly migratory species capable of long distance dispersal. Previous studies have suggested the presence of at least 4 demographically independent loggerhead turtle subpopulations nesting in the southeastern United States; however, these analyses were hampered by limited spatial coverage and small sample sizes in some cases.
We generated partial mtDNA control region sequences for 2,095 samples collected from 24 Atlantic coastal Florida beaches during the 2008 nesting and hatching season to supplement data
collected from the 2006 and 2007 seasons. Structure among rookeries was characterized using pairwise FST comparisons and analysis of molecular variance (AMOVA). Observed inter-annual
variation at some rookeries suggests the possibility of temporal variation due to differential cohort mixing. Regardless of temporal variation, a pattern of relatively consistent spatial
structuring is apparent. Haplotype frequencies may remain stable for over 150 beach kilometers. in some areas, but also may sharply transition across as little as 20 km with no apparent
impediment to female gene flow. The present study supports recognition of central and
southeastern Florida as distinct subpopulations. Further work is required using tools beyond
mtDNA to resolve the precise scale and patterns of rookery connectivity via female natal
dispersal.
Results: Genetic studies have provided invaluable information regarding population subdivision, particularly cryptic structure present in highly migratory species capable of long distance dispersal. Previous studies have suggested the presence of at least 4 demographically independent loggerhead turtle subpopulations nesting in the southeastern United States; however, these analyses were hampered by limited spatial coverage and small sample sizes in some cases.
We generated partial mtDNA control region sequences for 2,095 samples collected from 24 Atlantic coastal Florida beaches during the 2008 nesting and hatching season to supplement data
collected from the 2006 and 2007 seasons. Structure among rookeries was characterized using pairwise FST comparisons and analysis of molecular variance (AMOVA). Observed inter-annual
variation at some rookeries suggests the possibility of temporal variation due to differential cohort mixing. Regardless of temporal variation, a pattern of relatively consistent spatial
structuring is apparent. Haplotype frequencies may remain stable for over 150 beach kilometers. in some areas, but also may sharply transition across as little as 20 km with no apparent
impediment to female gene flow. The present study supports recognition of central and
southeastern Florida as distinct subpopulations. Further work is required using tools beyond
mtDNA to resolve the precise scale and patterns of rookery connectivity via female natal
dispersal.