Grant: 08-006R
Project Title: Genetic Population Structure of Green Turtles Nesting and Foraging in Florida
Project Manager: Campbell Nairn
Organization: University of Georgia Research Foundation - Natural Resources (Research and Educational Institute)
Grant Amount: $24,542.00
Completion Date: 2009-06-25
Summary: This research seeks to characterize the genetic diversity of green turtles nesting on Florida beaches and foraging in Florida waters. In collaboration with survey and tagging crews, samples representing nesting females have been collected from several Florida beaches during the 2007 nesting season. Utilizing these samples, we will use pairwise tests of mitochondrial haplotype frequency divergence to determine if sufficient structure exists to designate distinct management units among the rookeries comprising the Florida nesting aggregation. In collaboration with the UCF, the Inwater Research Group, and Quantum Resources/FPL, we will also examine the genetic structure of multiple green turtle foraging aggregations along Florida's Atlantic coast as well as the waters of Key West National Wildlife Refuge. This study will attempt to evaluate rookery contributions to each of the foraging grounds based on mitochondrial and microsatellite analyses. We will also screen for additional informative variation in the mitochondrial genomes of nesting green turtles representing key rookeries in the wider Caribbean with ultimate goal of developing haplotype-specific single nucleotide polymorphism (SNP) assays to improve the resolution of mixed stock analyses.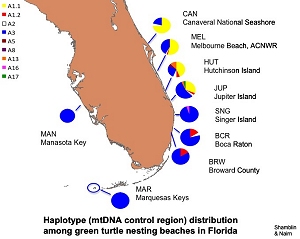 Results: A mtDNA haplotype database was developed for green turtles from Florida nesting beaches. Preliminary evaluation for management unit boundaries suggests that there are at least two subpopulations in Florida. Haplotype data for 417 individuals, representing 5 foraging sites, indicate protential source rookeries, but extensive haplotype sharing between nesting aggregations prevents precise estimations of contributions from individual sites and demonstrates the need to identify additional mitochondrial genome variation for mixed stock analyses. We have succeeded in identifying potentially informative variation for three rookery comparisons (Mexico vs. Florida Cm-A1.1, Mexico vs. Florida Cm-A1.2, Tortuguero vs. Suriname CmA5). (Author: Dr. Campbell Nairn)
Results: A mtDNA haplotype database was developed for green turtles from Florida nesting beaches. Preliminary evaluation for management unit boundaries suggests that there are at least two subpopulations in Florida. Haplotype data for 417 individuals, representing 5 foraging sites, indicate protential source rookeries, but extensive haplotype sharing between nesting aggregations prevents precise estimations of contributions from individual sites and demonstrates the need to identify additional mitochondrial genome variation for mixed stock analyses. We have succeeded in identifying potentially informative variation for three rookery comparisons (Mexico vs. Florida Cm-A1.1, Mexico vs. Florida Cm-A1.2, Tortuguero vs. Suriname CmA5). (Author: Dr. Campbell Nairn)