Grant: 09-011R
Project Title: Genetic Population Structure of Green Turtles Nesting and Foraging in Florida: Phase II. Management Unit Designations and Inter-annual Recruitment
Project Manager: Campbell Nairn
Organization: University of Georgia Research Foundation - Natural Resources (Research and Educational Institute)
Grant Amount: $34,321.00
Completion Date: 2010-09-23
Summary: This project will characterize the genetic diversity of green turtles nesting on Florida beaches and foraging in Florida waters. Based on preliminary results of the population structure survey of samples collected in 2007, collaborators increased sampling effort in 2008 to allow tests of intra-seasonal temporal variation at some sites and to clarify ambiguous relationships among many central Atlantic Florida nesting beaches. Analysis of the larger 2008 sample cohorts is critical to making robust inferences of management unit status as well as providing unbiased baseline data for mixed stock analyses. Mitochondrial sequencing of Florida nesting Cm-A1 and Cm-A3 individuals is over 50% complete, and comparisons between nesting beach and foraging samples suggest that we have identified at least one informative variable position that may diagnostically distinguish Florida and Mexican nesting Cm-A1s. Collaborators have made nesting beach samples from Tortuguero, Aves, and Suriname available for additional testing. We will complete initial screening by sequencing the majority of the mitochondrial genomes of at least 1 individual representing each major haplotype from each of the wider Caribbean rookeries with the aim of finding diagnostic variable positions. Funding will support performing mixed stock analyses on pelagic juvenile samples collected by Blair Witherington and annual cohorts of neritic juvenile samples collected at the St. Lucie Power Plant. The former represent the first pelagic green turtle samples available, and the latter provide an unparalleled opportunity to explore differential annual recruitment of juvenile green turtles in Florida waters.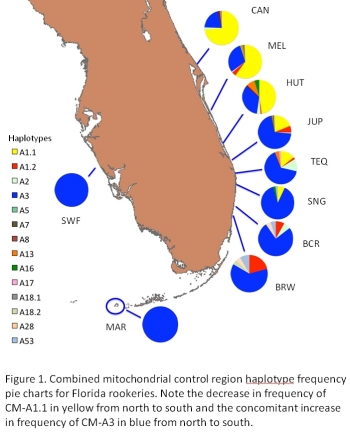 Results: Genetic studies have provided invaluable information regarding population subdivision,
particularly cryptic structure present in highly migratory species capable of long distance
dispersal. Previous studies have suggested the presence of at least 4 demographically
independent loggerhead turtle subpopulations nesting in the southeastern United States; however,these analyses were hampered by limited spatial coverage and small sample sizes in some cases.
Results: Genetic studies have provided invaluable information regarding population subdivision,
particularly cryptic structure present in highly migratory species capable of long distance
dispersal. Previous studies have suggested the presence of at least 4 demographically
independent loggerhead turtle subpopulations nesting in the southeastern United States; however,these analyses were hampered by limited spatial coverage and small sample sizes in some cases.
We generated partial mtDNA control region sequences for 2,095 samples collected from 24
Atlantic coastal Florida beaches during the 2008 nesting and hatching season to supplement data
collected from the 2006 and 2007 seasons. Structure among rookeries was characterized using
pairwise FST comparisons and analysis of molecular variance (AMOVA). Observed inter-annual
variation at some rookeries suggests the possibility of temporal variation due to differential cohort mixing. Regardless of temporal variation, a pattern of relatively consistent spatial structuring is apparent. Haplotype frequencies may remain stable for over 150 beach kilometers in some areas, but also may sharply transition across as little as 20 km with no apparent impediment to female gene flow. The present study supports recognition of central and
southeastern Florida as distinct subpopulations. Further work is required using tools beyond
mtDNA to resolve the precise scale and patterns of rookery connectivity via female natal
dispersal.